The Sennet Consortium sites develop innovative technologies that accelerate our understanding of cellular senescence. Our tools span imaging, spatial biology, and computational analytics, empowering researchers to uncover the complex roles senescent cells play in aging and disease.
Jason Ji, PhD (Duke University) and Anthony Qu (University of Pennsylvania) discuss and demonstrate “DeepScence,” an AI-powered senescence research program.
Dr. Jun Hee Lee (University of Michigan) discusses the development and use of seq-scope and FICTURE technology, and how they further the field of spatial transcriptomics, especially in regards to the study of senescent cells.
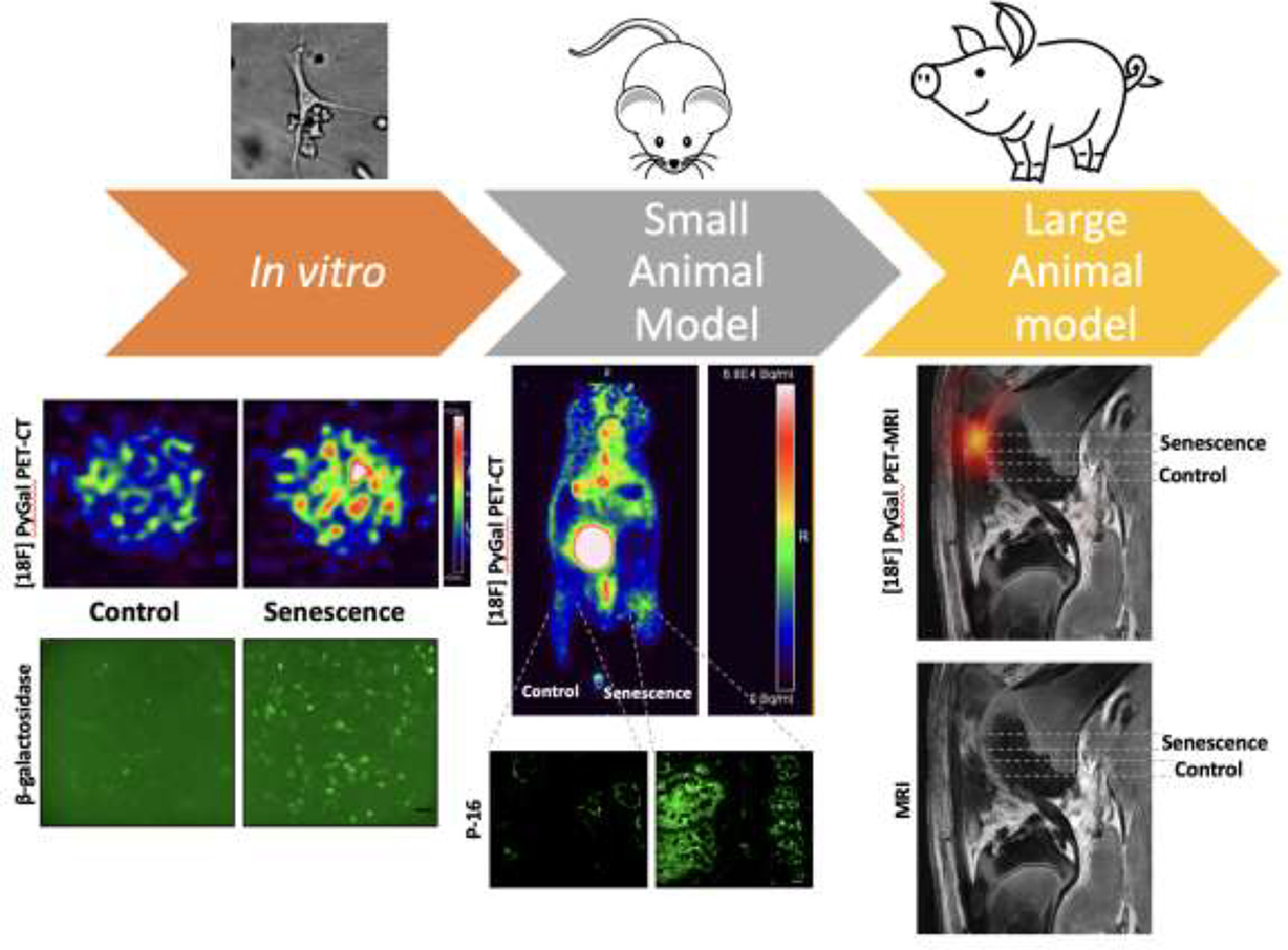
18F-PyGal Radiotracer
Institution: Stanford University
A groundbreaking radiotracer designed for in vivo detection of β-galactosidase activity, a hallmark of senescence. Using integrated PET/MRI imaging, 18F-PyGal enables precise, non-invasive visualization of senescent cells in live organisms, revolutionizing how researchers monitor cellular aging in real time.
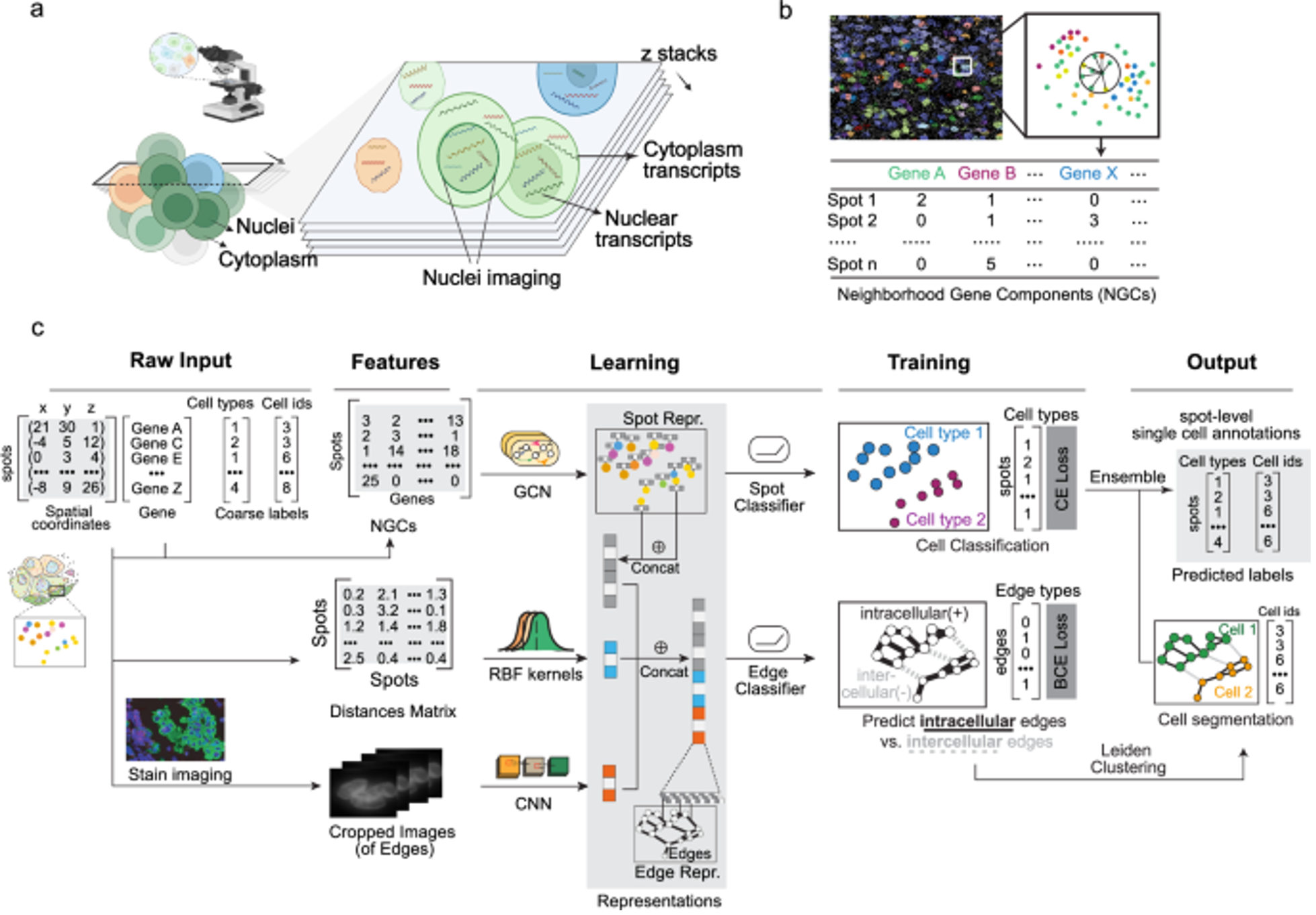
Bering
Institution: Massachusetts Institute of Technology
Bering is a joint cell segmentation and annotation tool designed specifically for spatial transcriptomics. It uses graph-embedding transfer to improve cell boundary detection and label assignment even in complex tissues. The method enhances downstream spatial analyses by producing more accurate cellular maps.
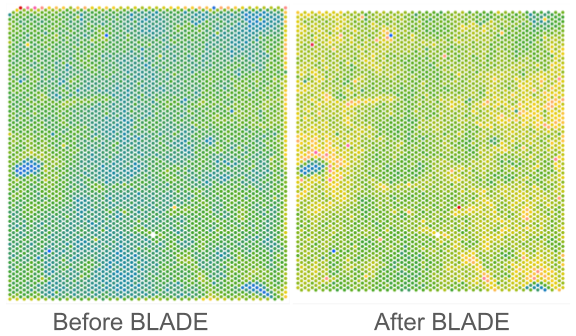
Border, Location, and edge Artifact DEtection (BLADE)
Institution: University of Minnesota
BLADE is a novel collection of automated cross-platform statistical methods for detecting and removing three commonly occurring artifacts in spatial transcriptomics data: (i) border effects, where total gene reads is modified at the border of the capture area; (ii) tissue edge effects, where total gene reads is modified at the edge of the tissue; (iii) location batch malfunctions, where there is a zone in the same location on all slides in a batch with substantially decreased sequencing depth. To date, BLADE has been successfully applied to both Visium and CosMx data, and can be extended to other spatial omics platforms as well.
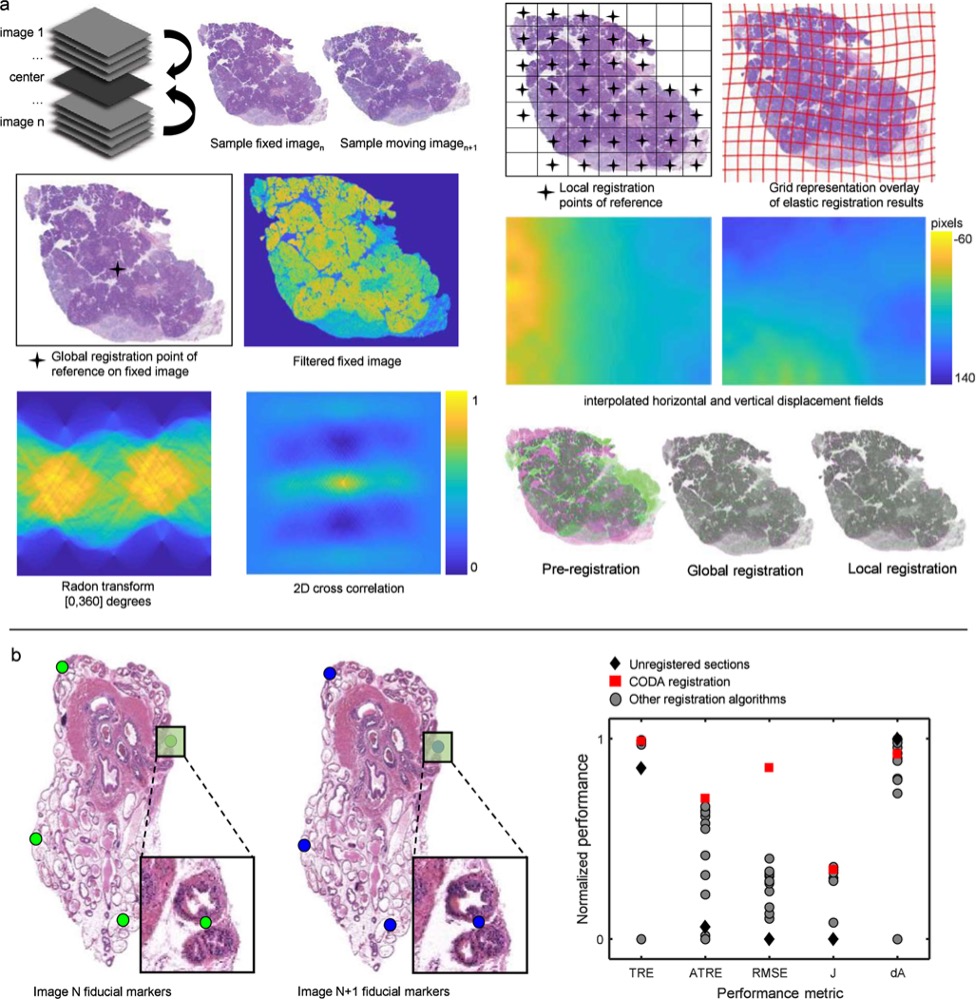
CODA
Institution: Johns Hopkins University
CODA reconstructs high-density image datasets into large-scale 3D tissue maps at single-cell resolution. Its computational pipeline integrates serial microscopy sections into coherent 3D volumes, enabling quantitative exploration of cellular architecture across centimeters of tissue. This approach supports structural and spatial studies at unprecedented scale.
DeepScence
Institution: Duke University
Accurately identifying senescent cells is essential for studying their spatial and molecular features. DeepScence, a method based on deep neural networks, identifies senescent cells in single-cell and spatial transcriptomics data. DeepScence is based on CoreScence, a senescence-associated gene set curated to incorporate information from multiple published gene sets. DeepScence can accurately identify senescent cells in single-cell gene expression data collected both in vitro and in vivo, as well as in spatial transcriptomics data generated by different platforms, substantially outperforming existing methods.
FICTURE
Institution: University of Michigan
Segmentation challenges in senescence research are a thing of the past. FICTURE, published in Nature Methods (2024), leverages deep learning to resolve complex tissue segmentation issues, dramatically improving the accuracy of cellular identification and quantification in histological images.
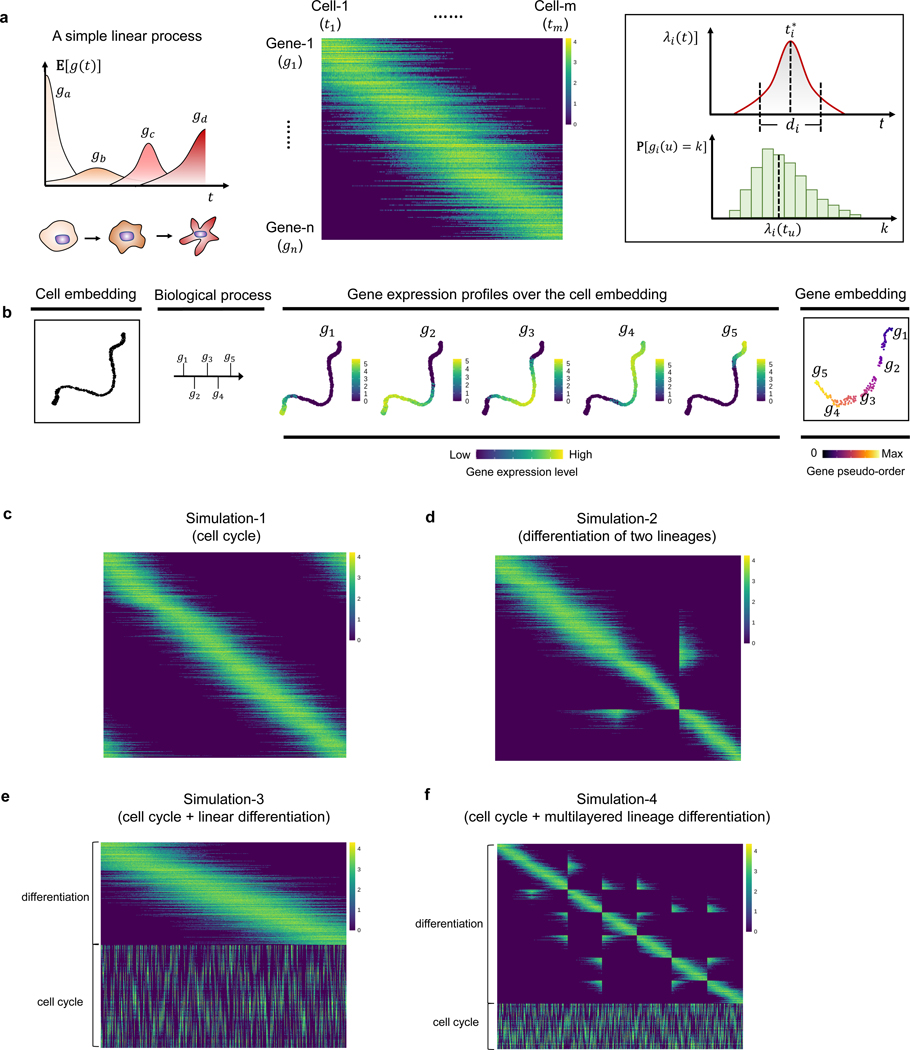
Gene Trajectory
Institution: Yale University
Gene Trajectory provides an alternative to pseudotime for modeling the evolution of senescent cell phenotypes. Using optimal transport metrics, it reconstructs cellular transitions with higher stability and interpretability. The method is particularly useful for datasets with gradual or branching transcriptional changes.
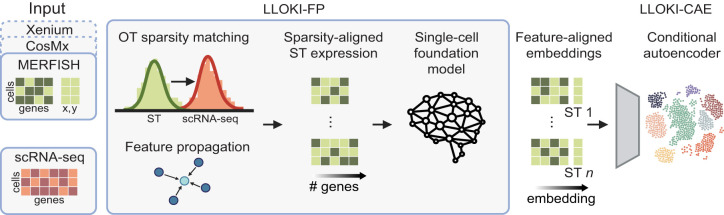
LLOKI
Institution: Brown University
LLOKI unifies spatial transcriptomics data from diverse platforms into a shared, coherent representation. It resolves differences in resolution, chemistry, and data structure to support integrated analyses. The framework enables cross-platform comparison and meta-analysis.
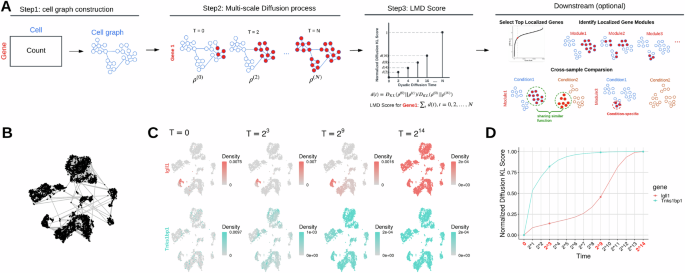
LMD
Institution: Yale University
The Localized Marker Detector (LMD) identifies gene expression signatures in locally clustered cells without relying on predefined cell clusters. Its multiscale approach detects subtle markers across heterogeneous populations. LMD enhances biological interpretation in complex or poorly separable tissues.
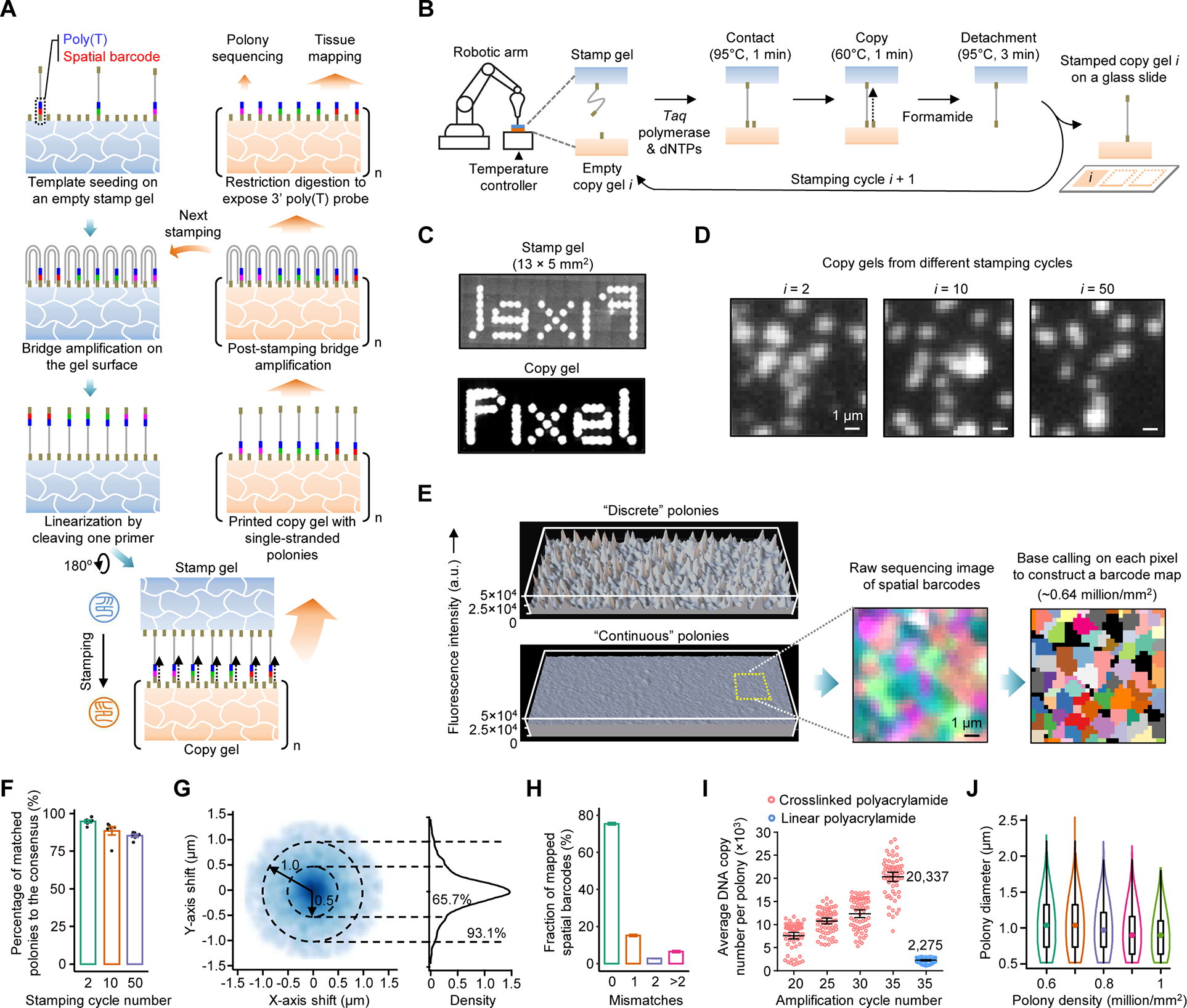
Pixel-Seq
Institution: University of Washington
An advanced single-cell spatial transcriptomic and proteomic assay, Pixel-Seq enables high-resolution mapping of senescent cells in complex tissues. Published in Cell (2022), this tool integrates RNA and protein data to provide a multidimensional view of senescent microenvironments.
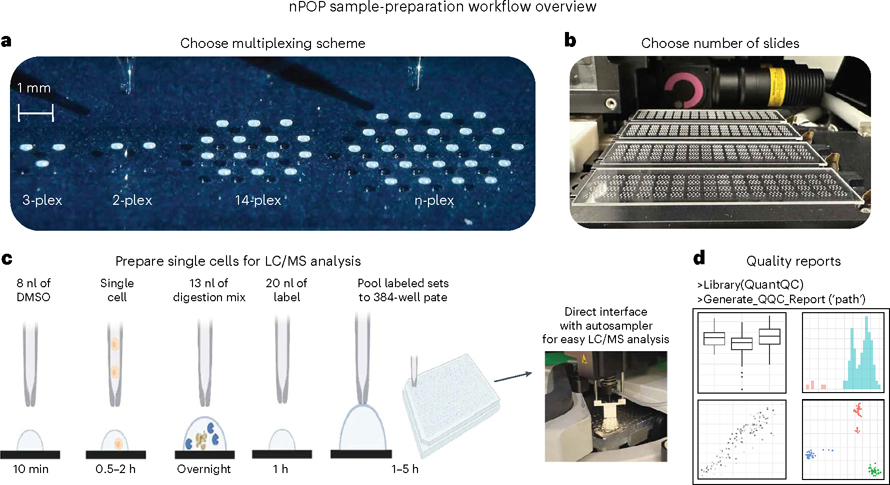
QuantQC
Institution: Massachusetts General Hospital
QuantQC is a quality control and correction toolkit for single-cell proteomics experiments, particularly those using nPOP sample preparation. It identifies and corrects protein leakage and other technical artifacts to improve quantification fidelity. The tool strengthens reliability across large-scale proteomic datasets.
ScResolve
Institution: Carnegie Mellon University
ScResolve recovers single-cell expression profiles from mixed-cell datasets, unlocking high-resolution insights from multicellular-level data. Featured in Cell Reports Methods (2024), this computational tool enables a deeper understanding of heterogeneous cell populations, particularly useful in tissues rich in senescent cells.
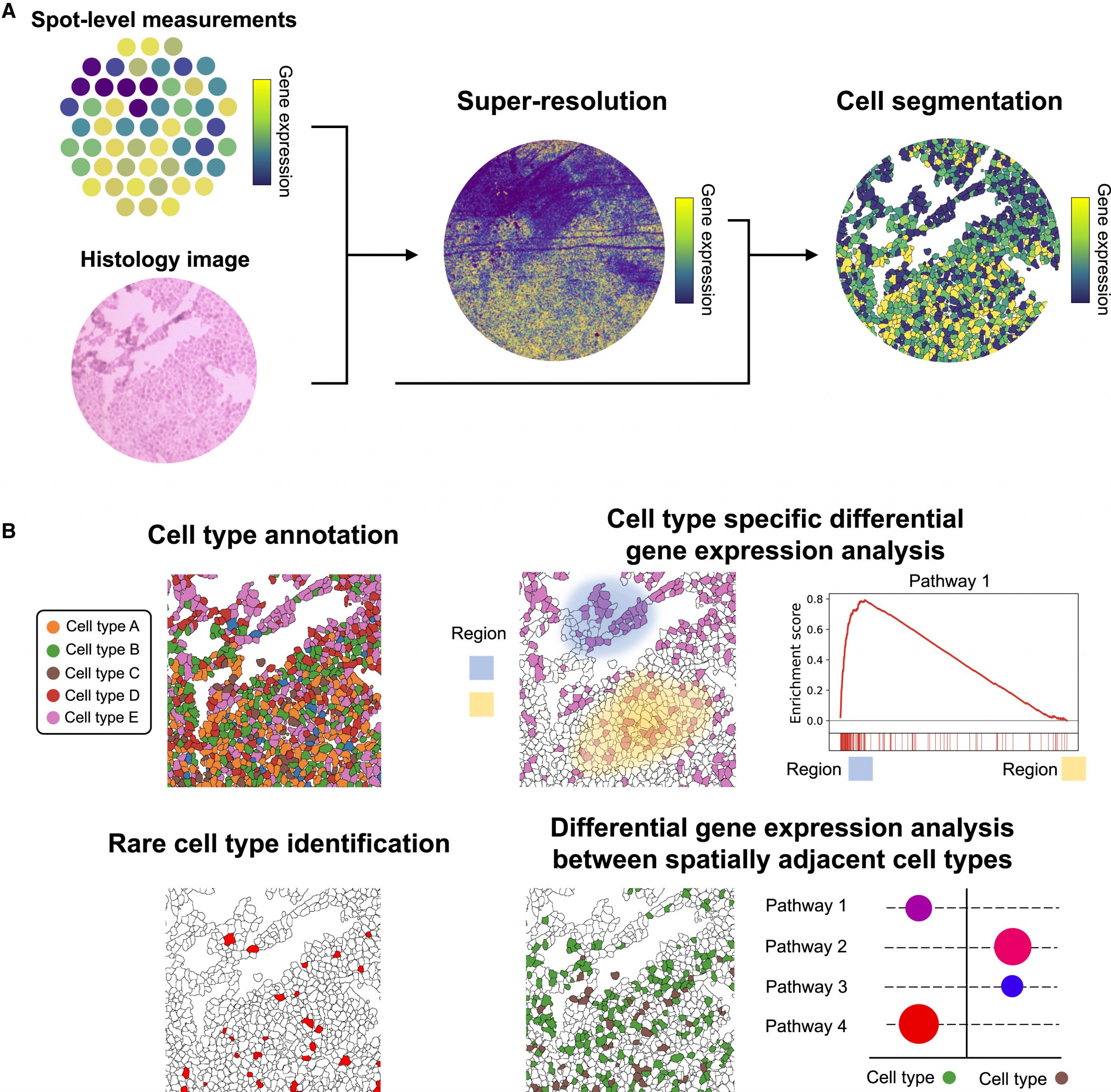
Sen Predictor
Institution: Buck Institute for Research on Aging
Sen Predictor uses deep learning applied to nuclear morphology to predict cellular senescence at high accuracy. Trained on fibroblast datasets, it generalizes to H&E-stained tissue sections. The method provides a fast, non-molecular senescence detection strategy.
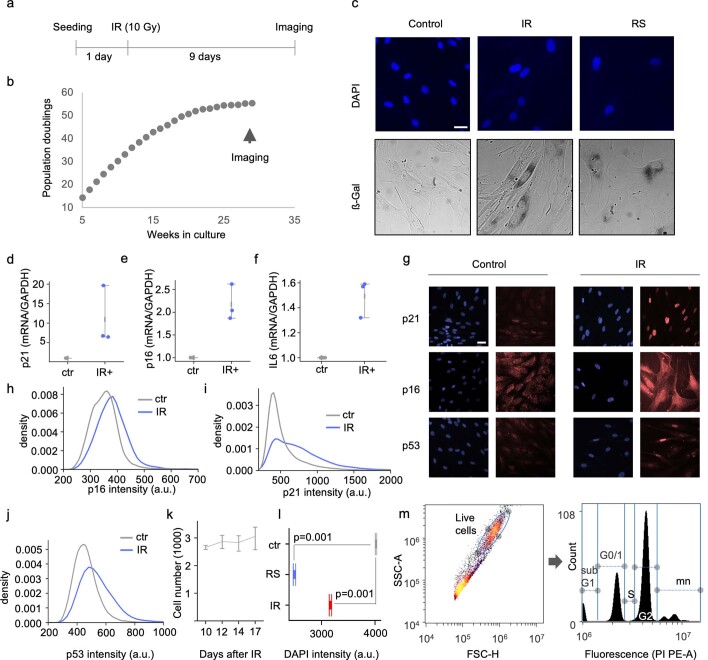
SenoQuant
Institution: Mayo Clinic
Fast, reliable, and user-friendly, SenoQuant is a precision image analysis platform that quantifies senescence biomarkers in human tissue. Whether analyzing large-scale cohorts or small biopsy samples, SenoQuant provides robust measurements critical for clinical and translational research.
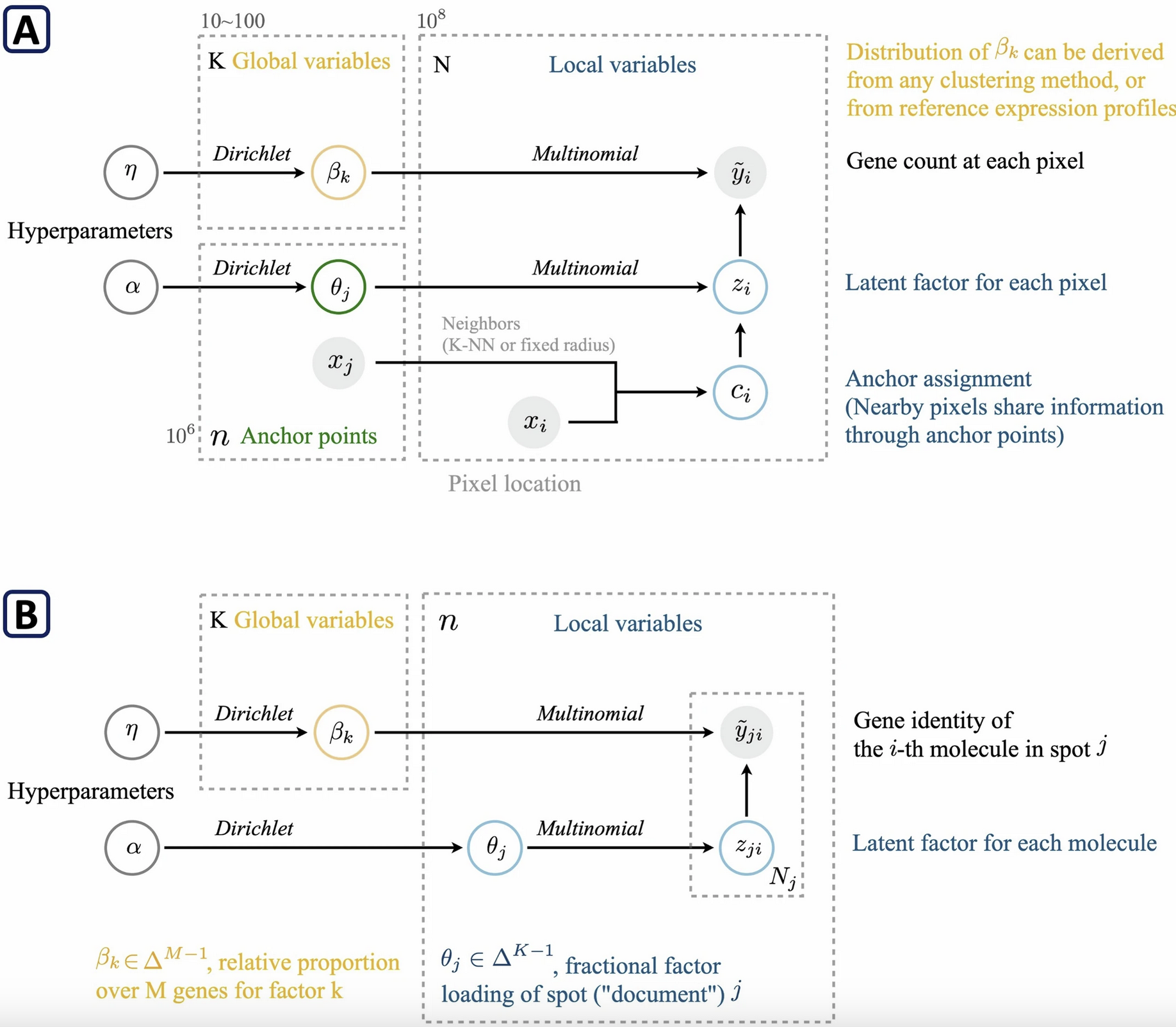
Seq-Scope
Institution: University of Michigan
Pushing the boundaries of spatial transcriptomics, Seq-Scope offers ultra-high resolution capabilities ideal for detecting rare and dispersed senescent cells. Published in Nature Protocols (2024), this technology provides an unprecedented spatial scale, allowing researchers to explore tissue architecture with single-cell clarity.
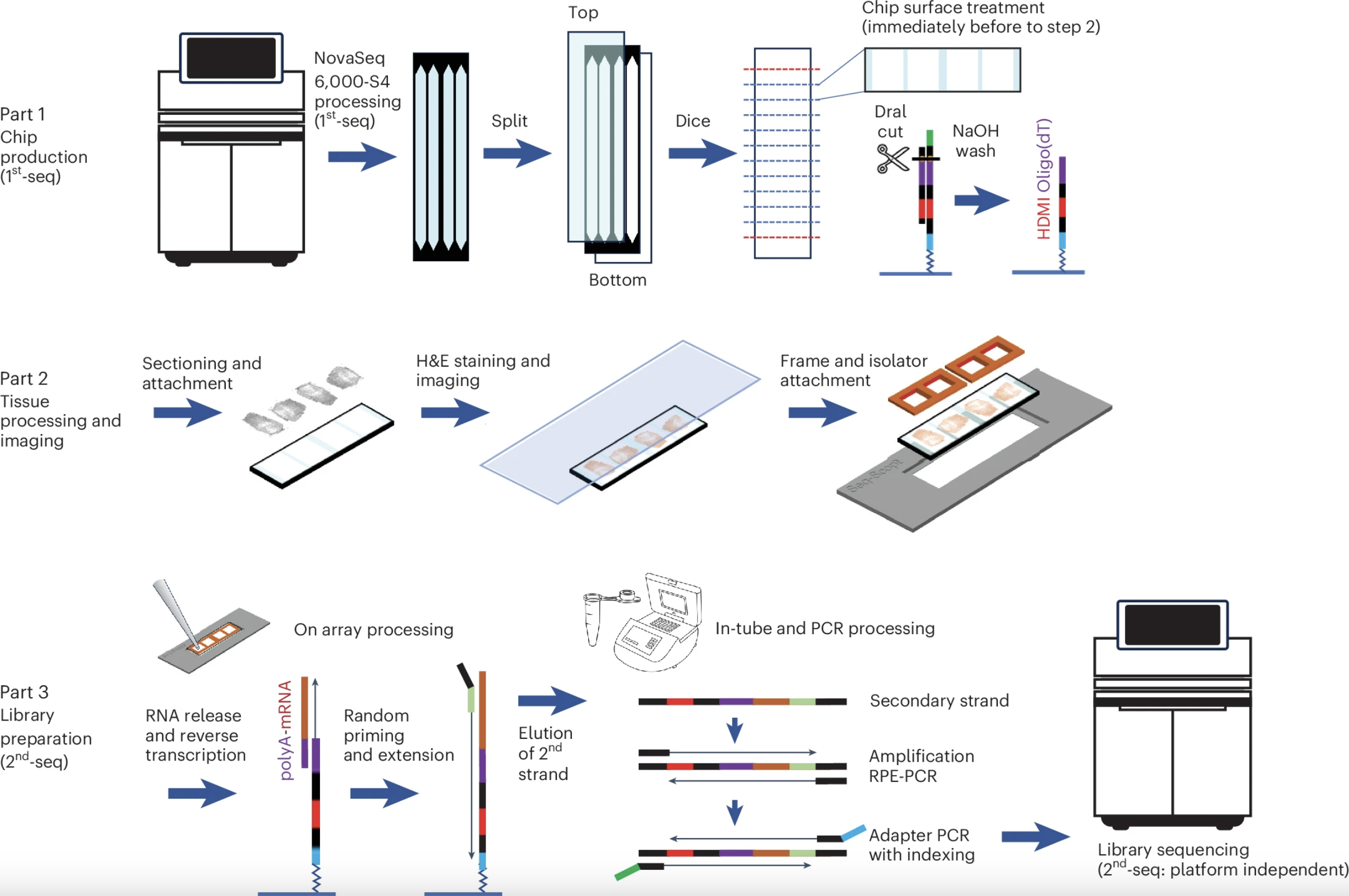
Seq-Scope-X
Institution: University of Michigan
Seq-Scope-X (Seq-Scope-eXpanded) combines the sub-micrometer spatial transcriptomic chemistry of Seq-Scope with tissue-expansion microscopy to achieve resolution beyond the optical diffraction limit. This approach enables high-content, subcellular transcriptomic and proteomic analysis while preserving tissue morphology—offering nanoscale insights into cellular architecture and function.

SIMVI
Institution: Yale University
SIMVI is a deep variational framework that separates intrinsic cellular features from spatially induced ones in spatial omics data. It enables annotation-free characterization of how microenvironmental context shapes cell states. The tool enhances interpretation of cell–cell interactions and tissue organization.
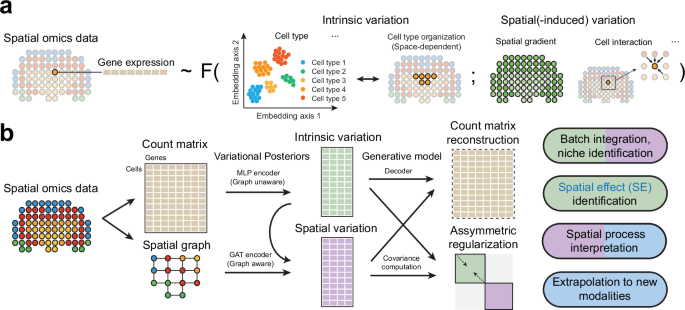
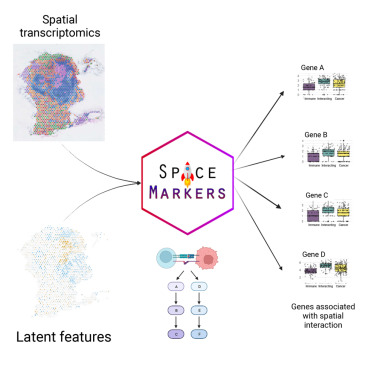
SpaceMarkers
Institution: Johns Hopkins University
SpaceMarkers identifies molecular changes driven by cell–cell interactions using latent space modeling. It detects spatially modulated gene programs without requiring explicit neighborhood definitions. The method helps dissect interaction-driven changes in senescent or tumor microenvironments.
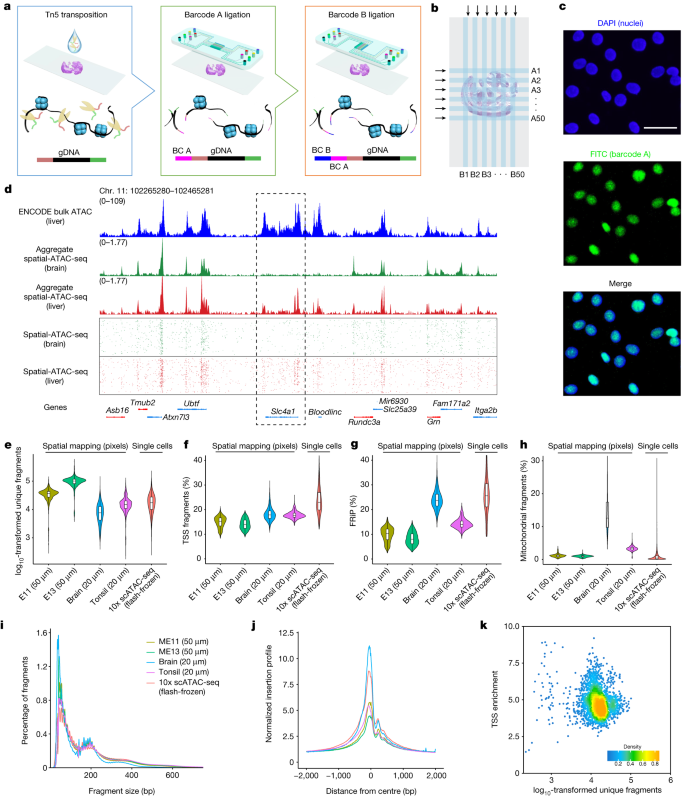
Spatial ATAC-seq
Institution: Yale University
Spatial ATAC-seq profiles chromatin accessibility directly in tissue sections, enabling spatially resolved epigenomic mapping. It reveals how local microenvironments shape regulatory landscapes. This method adds a crucial regulatory layer to spatial omics.
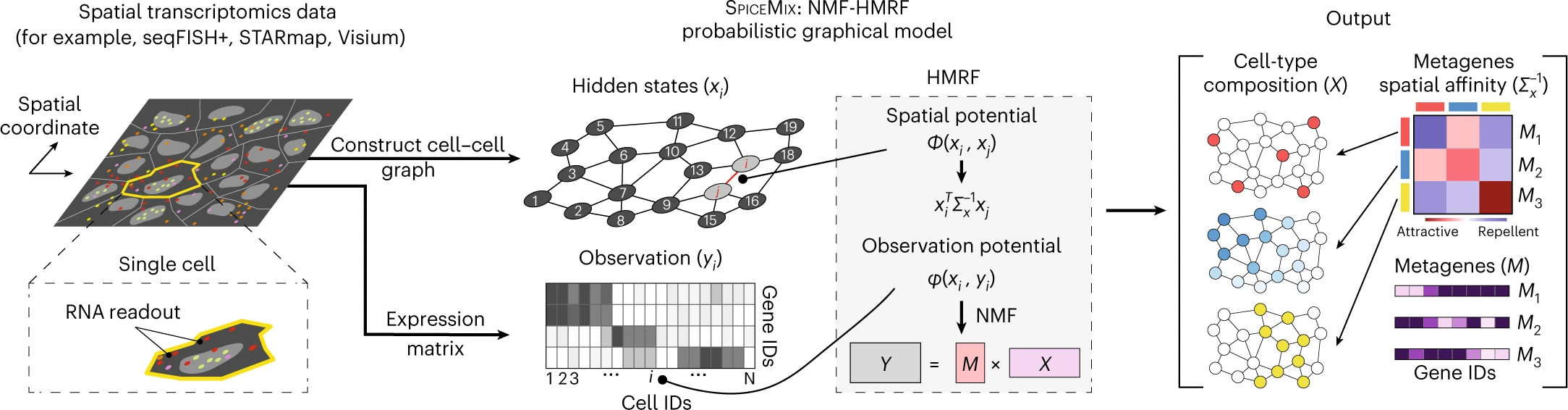
SPICEMIX
Institution: Brown University
SPICEMIX integrates spatial context with gene-expression profiles to infer latent cell states and microenvironmental structures. Its probabilistic model captures both molecular and spatial cues simultaneously. The tool enables robust identification of niche-specific phenotypes.